A new method to detect kinship in RNAseq data
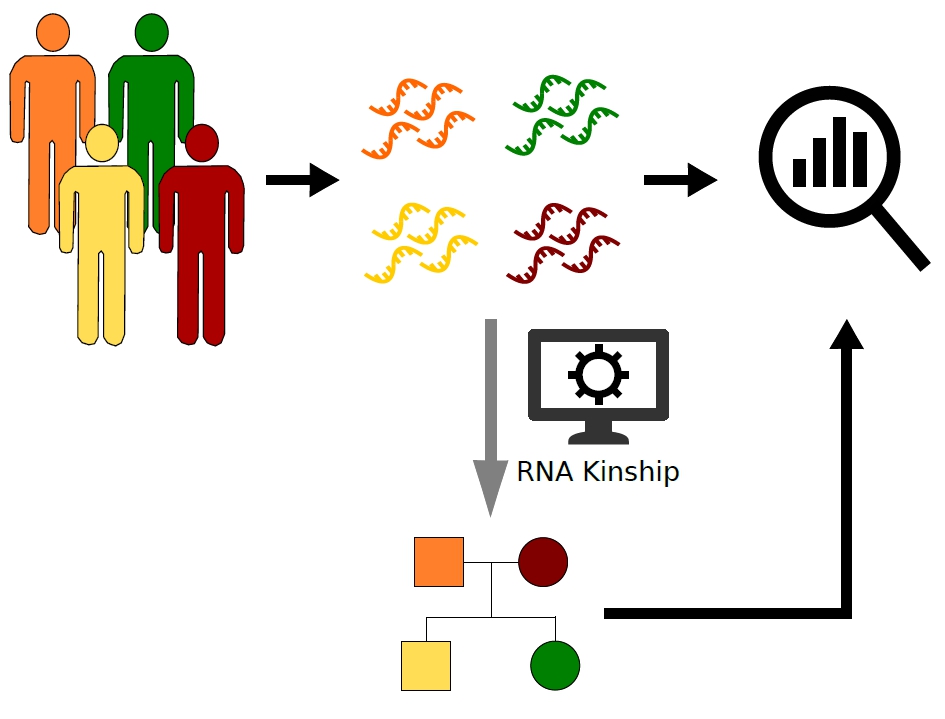
The Regulatory Genomics Group at the IGTP and the IJC has developed a new method to make checking whether samples that have had RNA sequenced are from related individuals. They have shown that it can be used on large data sets of RNA sequencing information to check if samples are from related individuals or not, or to reconstruct family trees.
Researchers can now access large amounts of data in scientific databases on genetic and associated information for large groups of individuals produced in previous studies, without having to generate the information themselves. The data is anonymized, but for many studies it is important to know whether individuals are related; in some cases when studying hereditary diseases in families, when they should be related, or in others when looking at large populations to discover patterns and trends, where the inclusion of related people will create a bias. Additionally, samples can be mislabelled; as the process from taking a sample, such as blood, extracting the genetic material, sequencing it and storing the data is a long chain of steps, each involving different people, with the associated risk of mistakes creeping in.
Finding the relationship between individual samples from genetic data (DNA) is possible to do. However, some studies only generate information on RNA, a similar molecule generated in cells but that can tell us more specific information about the health of a cell, or individual, and the importance of any mutations the person carries. . "Until now, researchers using RNA data had to also have access to DNA data to check whether samples were from related individuals," explains Natalia Blay, who carried out much of the work. "They had to download enormous files and carry out another whole layer of analysis; we wanted to know if we could reliably get this information from the RNA sequence data directly."
After developing a method to detect related individuals from RNA Sequence information, the team tested it in large group of samples from the GCAT Project, for whom the family relationships are well documented. Experts from the GCAT Project also contributed their expertise to this work.
"Using our technique we can successfully detect whether RNA samples are from related people and we can even construct family trees," says Tanya Vavouri, who led the study. "Researchers can confidently establish kinship for individuals in the many data sets for RNA sequencing data available. This means they do not have to spend time and computing power downloading and analysing associated genomic data, which doesn't always exist for all data sets. It means using RNA sequencing data is easier and cheaper in the long run."
Having access to the GCAT Project expertise has made this work easier to carry out.
"Our group have some experience in kinship analysis from DNA data," says Rafa de Cid, Director of the GCAT Project. "This has allowed us to collaborate with Tanya Vavouri's team to develop this method for RNA data," he added.
RNA sequencing data sets can be used to study a huge number of human diseases. This type of data mining is a valuable source of information for scientists studying individual diseases, their occurrence in populations or risk factors. This new methodology will help simplify this work for scientists. It is available free online for researchers who wish to use it.
Original paper
Assessment of kinship detection using RNA-seq data. Natalia Blay, Eduard Casas, Iván Galván-Femenía, Jan Graffelman, Rafael de Cid, Tanya Vavouri, Nucleic Acids Research, gkz776,
Published: 10 September 2019
Funding Information
This work was funded by the Spanish Ministry of Economy and Competitiveness [BFU2015-70581 and ADE 10/00026], by the Catalan Agency for Management of University and Research Grants [2017 SGR 1262 and 2017 SGR 529] and the CERCA Programme/Generalitat de Catalunya. Research at the IJC was supported by the "La Caixa" Foundation, Josep Carreras International Foundation and Celgene Spain. The Catalan Agency for Management of University and Research Grants (AGAUR) funded the open access grants.
The GCAT is an International project at the IGTP project carried out in collaboration with the Blood and Tissue Bank of Catalonia (BST) with the full support of the Directorate General of Research and Innovation and the Ministry of Health of the Generalitat of Catalonia. The initial phases of the project was supported by the Ministry of Health, Social Services and Equality of Spain and the Ministry of Health of the Generalitat of Catalonia via competitive public funding from the "Sub-Programme of Revitalizing Actions in the Research and Technology Environment" of the SNS.2010 (ADE10/00026, FIS).
